|
I am an Assistant Professor - Fixed Term in the Plant Research Laboratory, the Department of Plant Biology, and the Great Lakes Bioenergy Research Center (GLBRC) at Michigan State University (MSU). During my Ph.D. studies in the Department of Molecular Biosciences at the University of Texas at Austin, I mainly investigated the molecular mechanism of hybrid vigor, known as heterosis, in maize using an integrative approach of experimental biology and genomics. After completing my Ph.D. studies, as a postdoc, I joined the lab of C. Robin Buell at MSU for bioinformatics training. Following the successful training, I joined the lab of Federica Brandizzi where, as a postdoc, I investigated gene regulatory mechanisms underlying responses to endoplasmic reticulum stress in plants using systems-level approaches. Particularly, I have developed coexpression network-based gene discovery pipelines and applied them to pinpointing regulatory hub genes for functional characterization. In my current position, I investigate how genes are dynamically regulated in plants, in response to climate change using multi-omics network approaches. My research is funded by GLBRC through the Brandizzi Lab and by the MSU GREEEN project for which I serve as a PI./ / / / / Bio / Google Scholar |

|
|
I am a plant biologist driven by a passion for addressing critical biological questions at the systems level through a hypothesis-driven approach, harnessing the power of genomics. Just as individuals in society interact, so too do genes and proteins within cells. But what are the functional consequences of these molecular interactions in regulating biological pathways? These interactions, known as “biological networks,” are essential for maintaining cellular homeostasis in all living organisms. My long-term research goal is to unravel these gene networks and apply the insights to advance translational research. Representative papers are highlighted in yellow below. |
|
|
|
Adhikari B, Verchot J*, Brandizzi F, Ko DK* *Co-corresponding authors J Biol Chem. 2025 Feb 25;301(4):108354. Description: Over the past decade, our understanding of how the plant UPR machinery engages with pathogen infections, especially viral infection, and the functional consequences has greatly advanced because of genetic, genomic, and technological innovations as well as the accumulation of new knowledge of plant UPR. In this review, we provide a comprehensive overview of plant UPR signaling highlighting recent advances that reveal how it is tuned to orchestrate interconnected signaling pathways, thus operating as a moderator to control cell fate. |
|
|
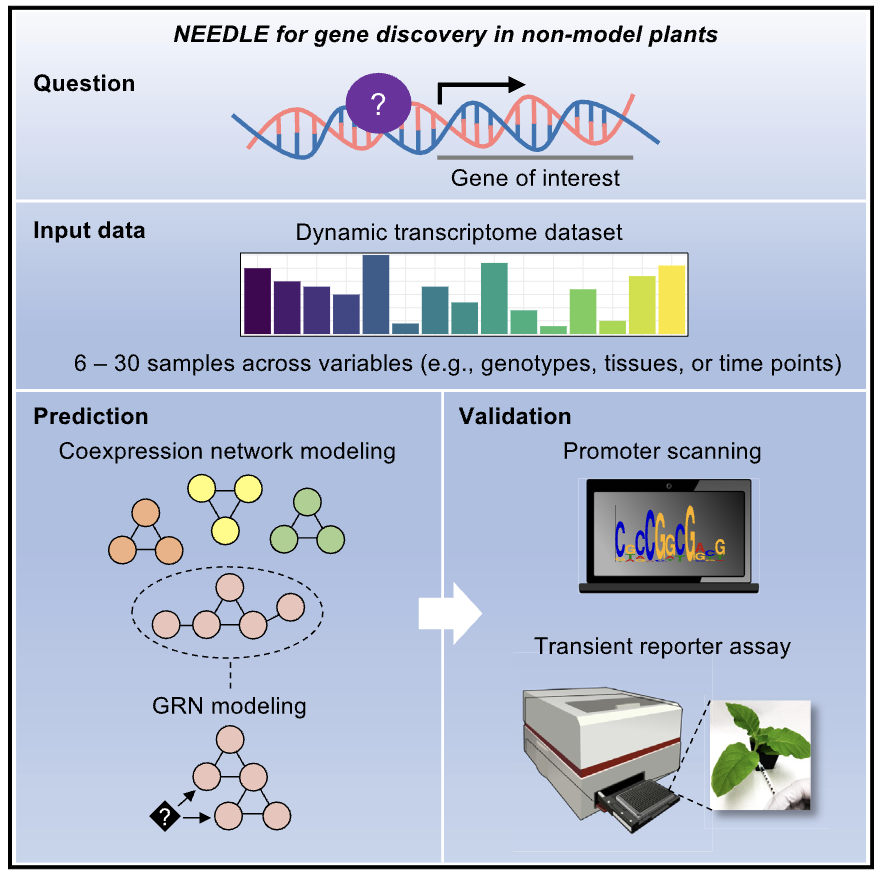
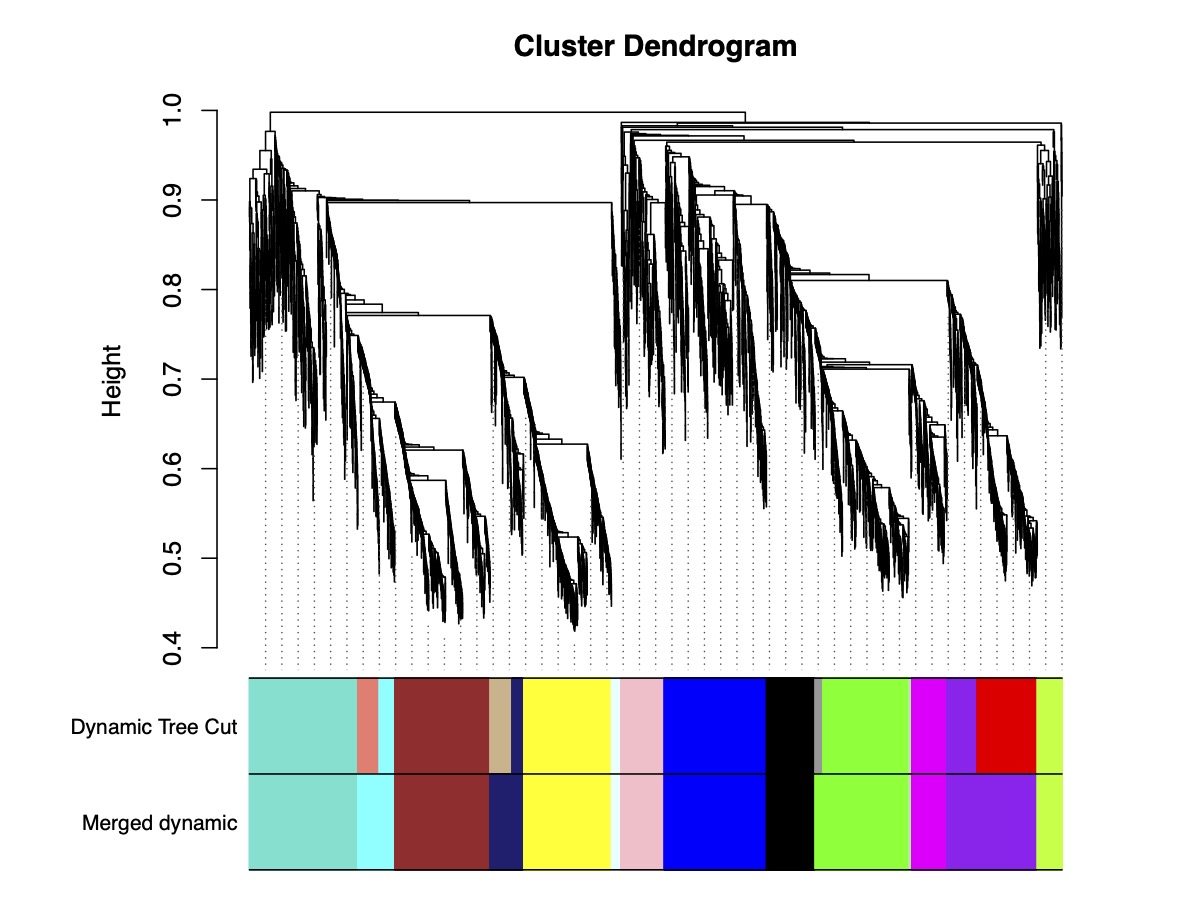
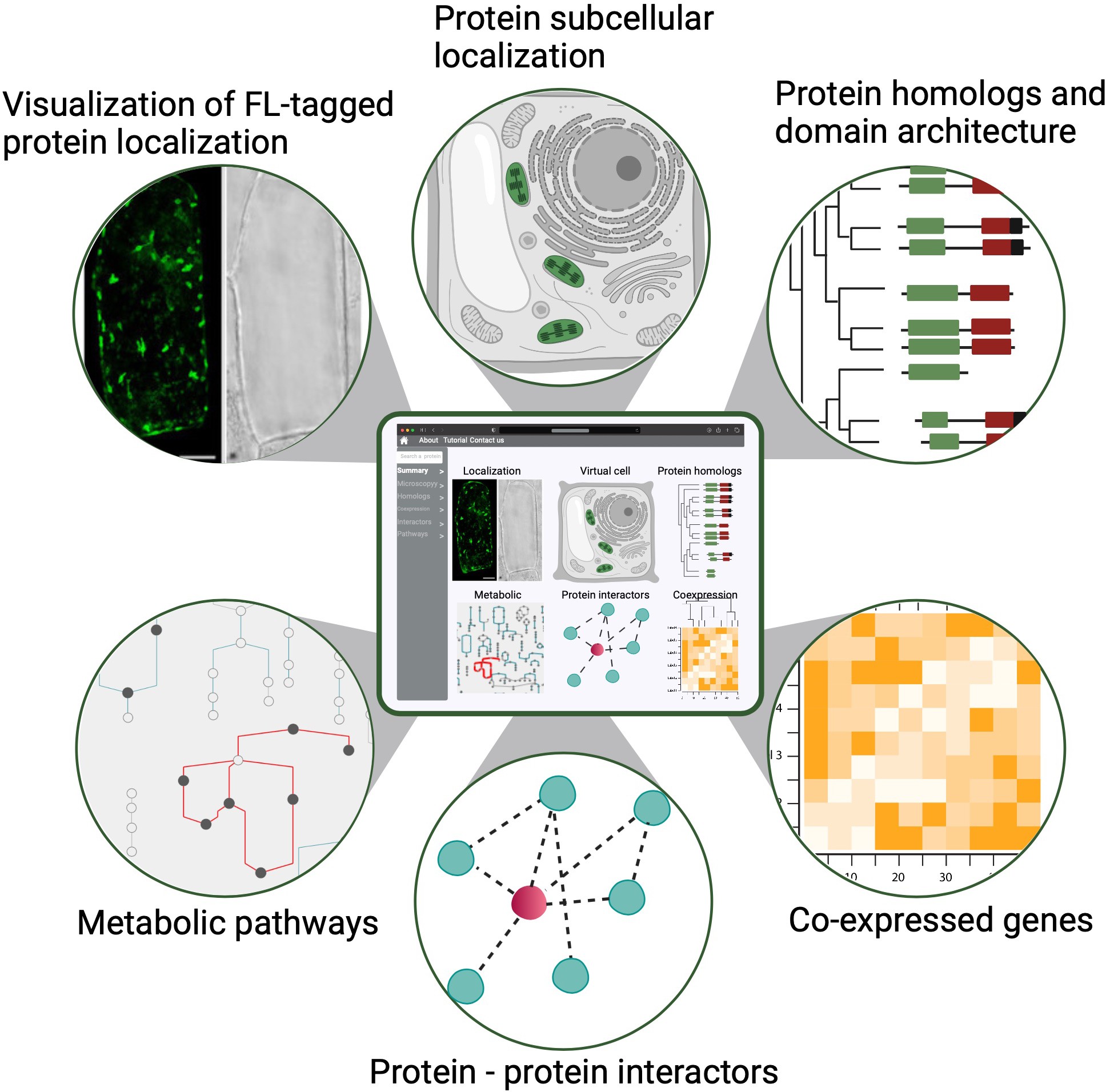
Ko DK, Brandizzi F Cell Rep Methods. 2025 Jan 27;5(1):100963. Highlighted in MSU-PRL bulletin / Highlighted in GLBRC bulletin / Github page for scripts Description: Meeting the energy and food demands of a growing global population demands the swift development of climate-resilient crops, achievable through targeted gene reprogramming for growth, development, and stress responses. Accurate prediction of transcription regulators is crucial for accelerating crop improvement via targeted engineering and breeding approaches. However, this is constrained by the scarcity of multi-omics datasets for most non-model species. To address this challenge, we present a robust network-based gene discovery pipeline that identifies transcription factors upstream of genes of interest by leveraging transcriptomic dynamics in non-model plants followed by rapid in planta experimental validation. |
|
|
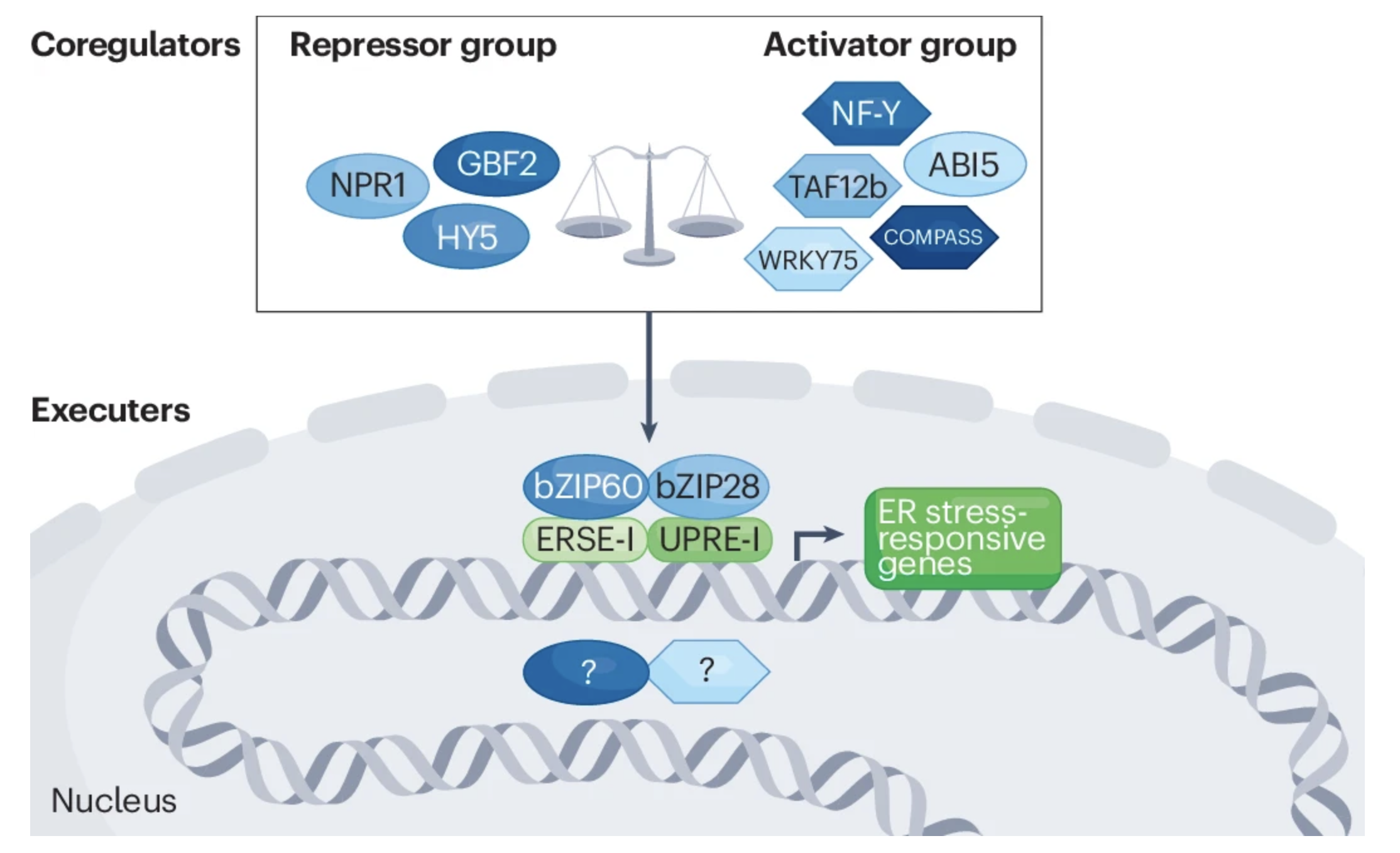
Ko DK, Brandizzi F Nat Rev Genet. 2024 Jul;25(7):513-525 Description: Plants have uniquely adapted to manage endoplasmic reticulum stress triggered by protein misfolding. We review the dynamics of gene expression regulation underlying the unfolded protein response in plants, highlighting recent insights provided by systems-level approaches and omics data. |
|
|
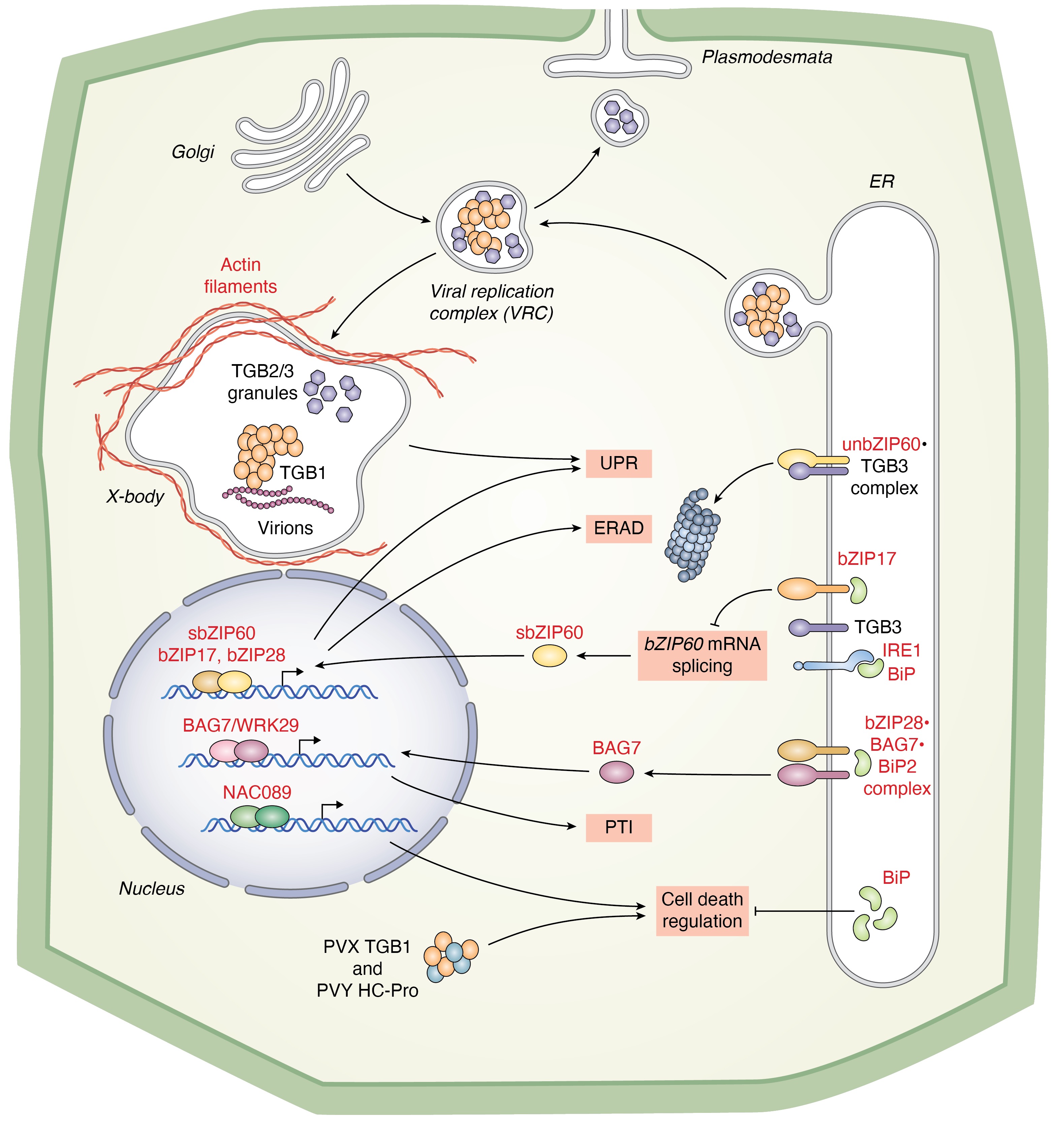
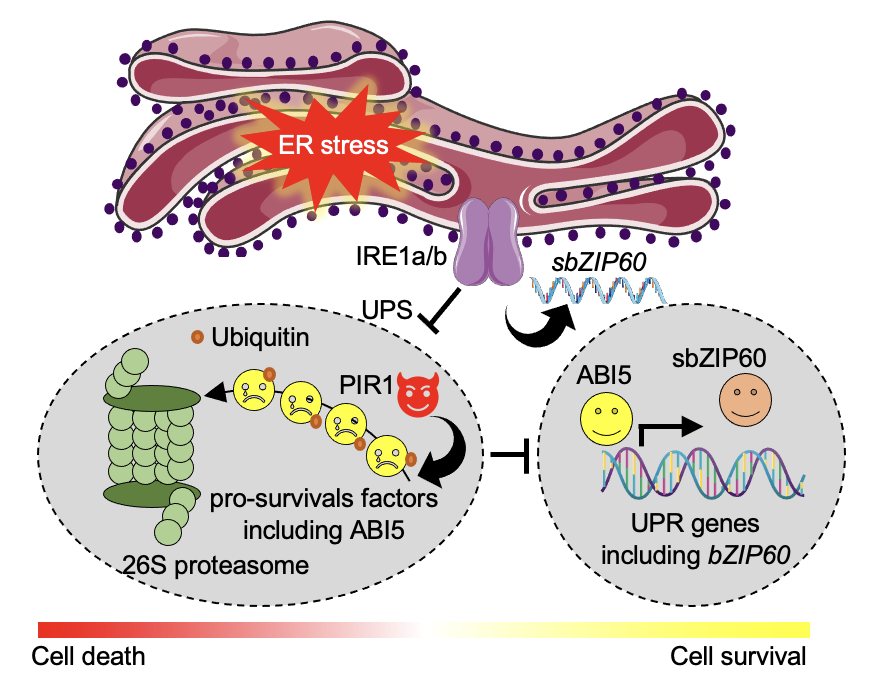
Ko DK, Kim JY, Thibault ET, Brandizzi F Nat Plants. 2023 9:1333-1346. Spotlight by Varshney et al. Trends Plant Sci / Github page for scripts / Media highlight Description: In this study, we performed a forward genetic screening and discovered a new regulator that helps cells determine life or death under unresolved ER stress. Using an interdisciplinary approach of omics, biochemistry, and genetics, we identified its working mechanism and functional relationship with existing regulators. |
|
|
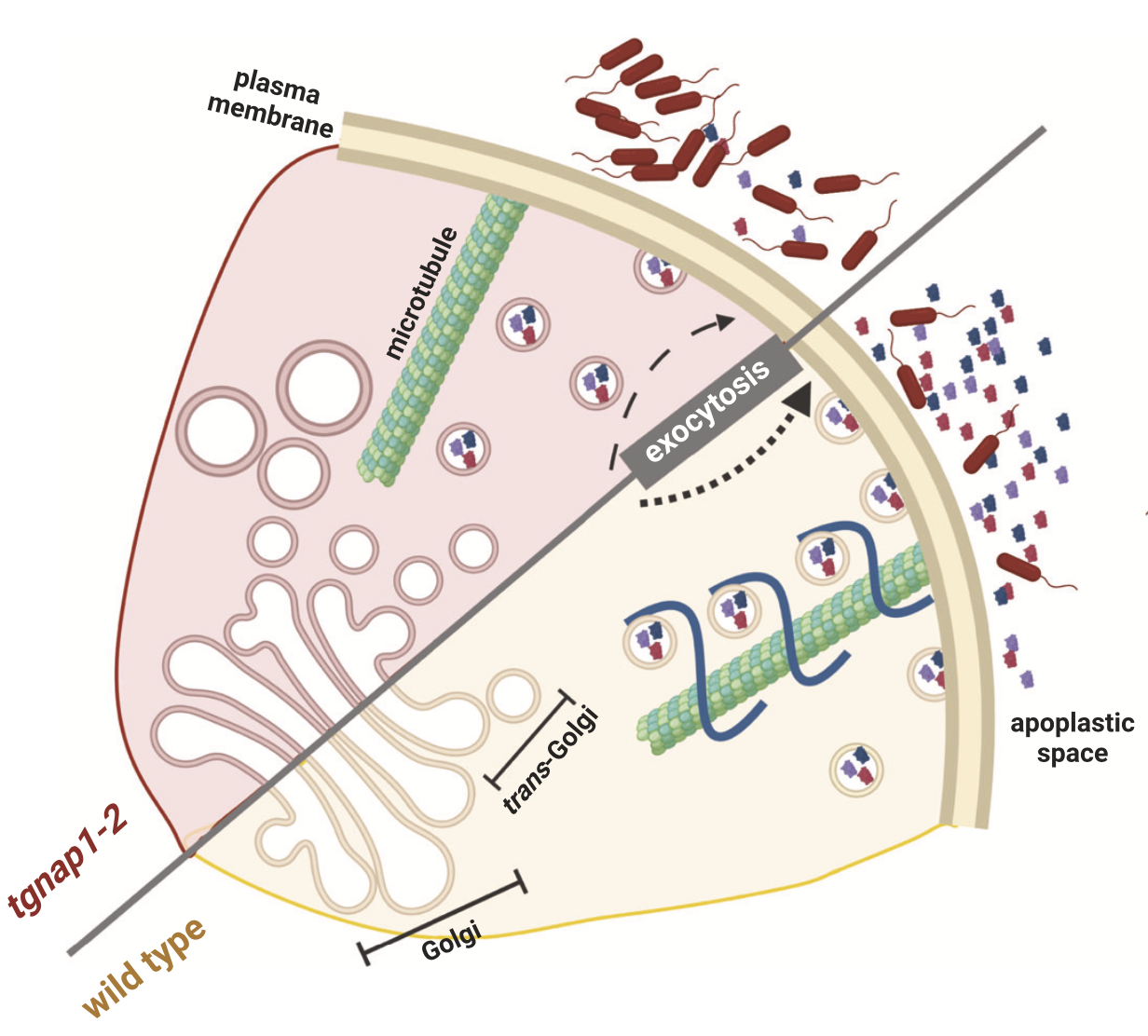
Bhandari BD, Ko DK, Kim SJ, Nomura K, He SY, Brandizzi F Nat Commun. 2023 14,6357. Description: Defining plant defense machinery against pathogens is significant in cell biology and crop yield. This paper shows that TGNap1, a TGN and microtubule-binding protein, is required for defense and efficient anti-microbial protein secretion, linking secretion and cytoskeleton. I performed the RNA-seq analysis as a co-author. |
|
|
Ko DK, Brandizzi F Plant Proteostasis. Methods in Molecular Biology, 2023 vol 2581. Humana, New York, NY Github page Description: The paper provides a step-by-step workflow for coexpression network analyses using WGCNA and discusses its potential applications. |
|
|
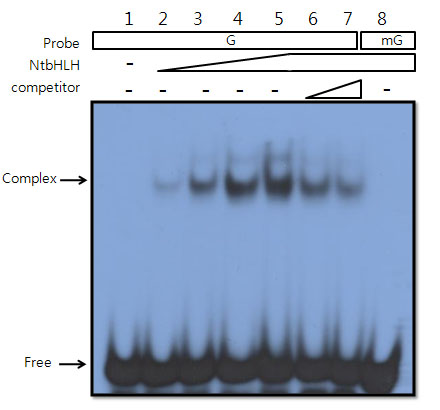
Ko DK, Brandizzi F Nat Plants. 2022 May;8(5):481-490. Github page for scripts / Media highlight Description: We discovered a new UPR regulator and characterized its functional role in the UPR at the mechanistic level through a multi-omics approach. |
|
|
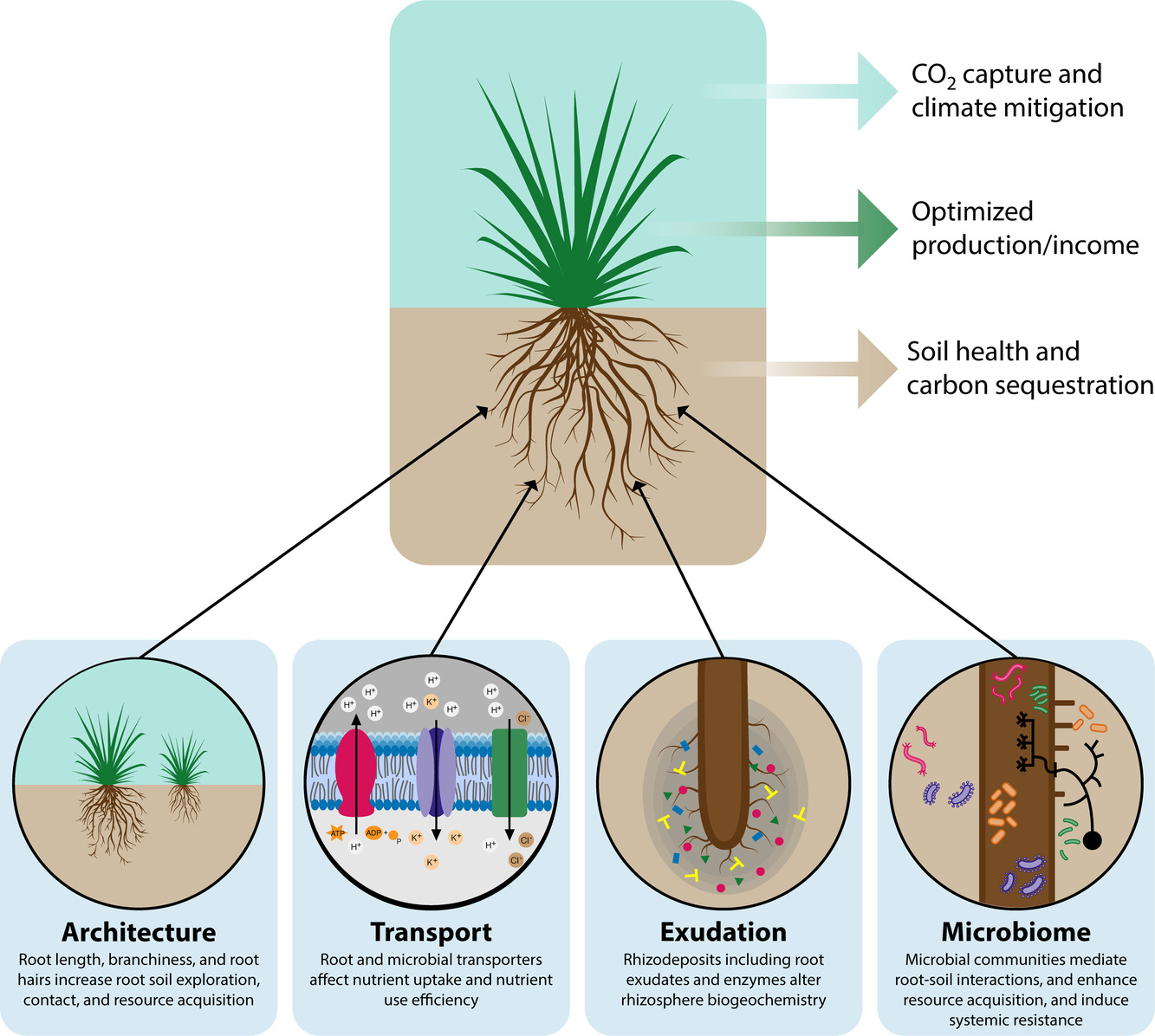
York LM, Cumming JR, Trusiak A .... Ko DK .... Yang WH The Plant Phenome Journal 5.1 2022: e20028. GLBRC website Description: In this white paper, the GLBRC colleagues and I discussed the significance and challenges of root biology in bioenergy research. |
|
|
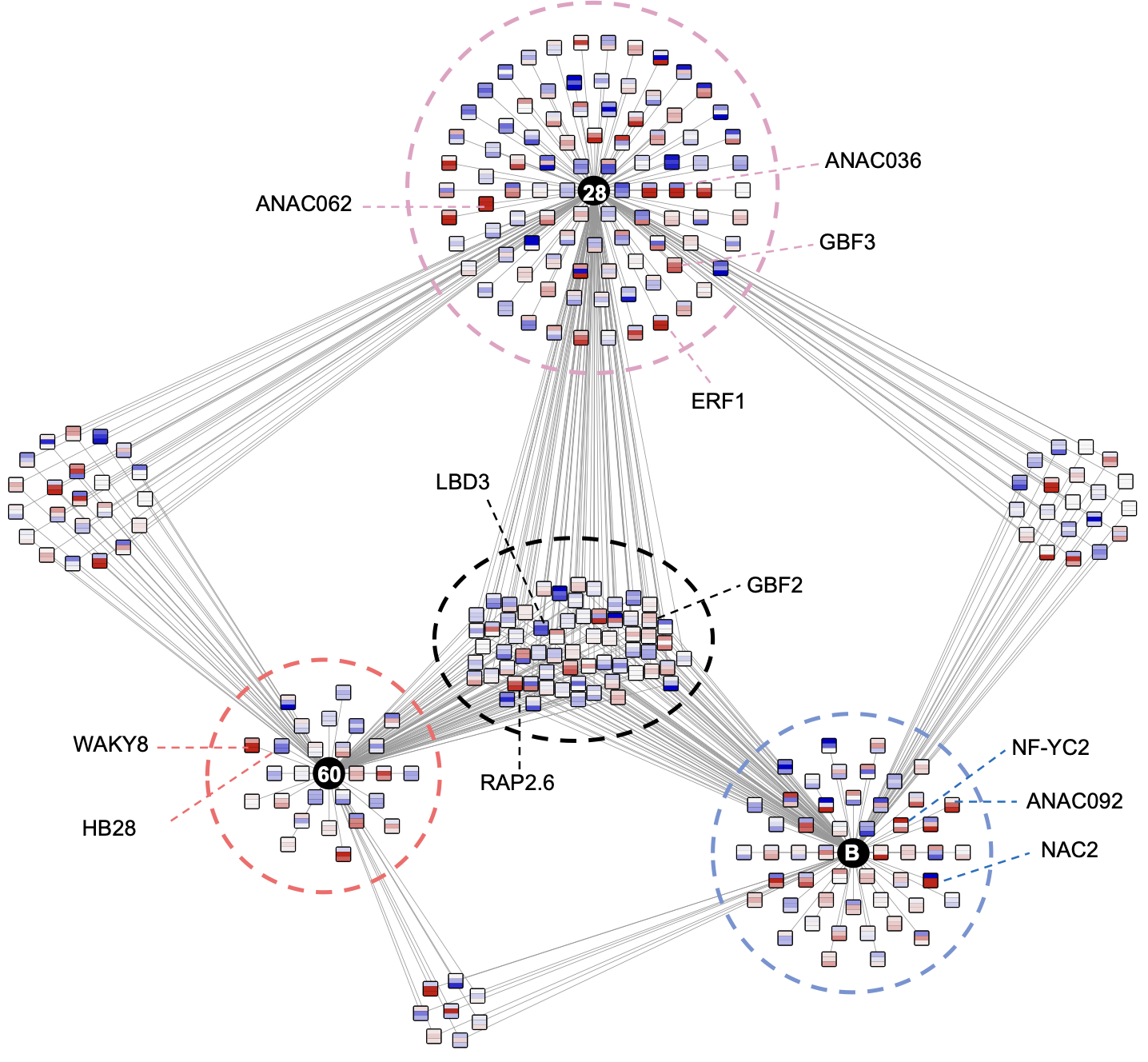
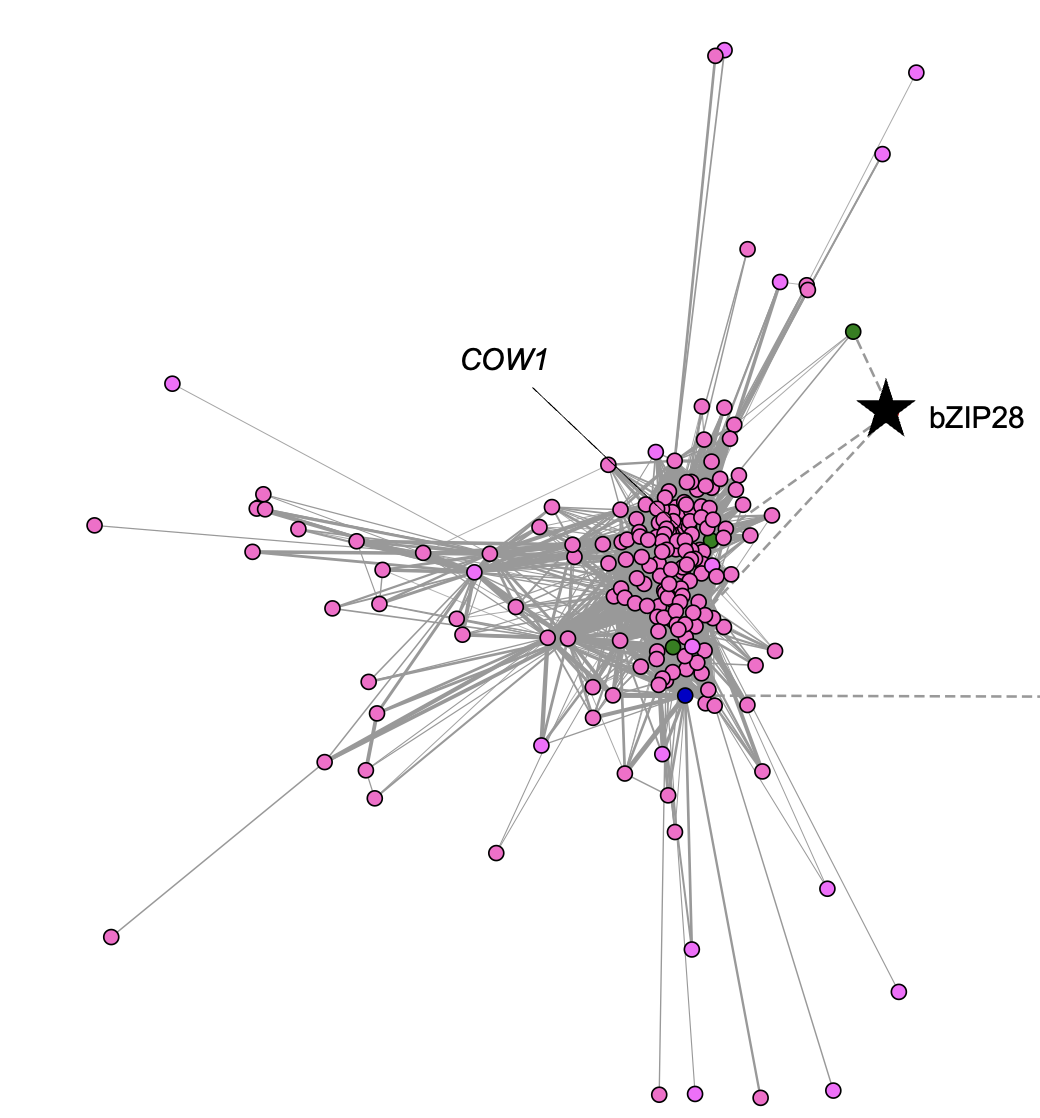
Ko DK, Brandizzi F Commun Biol. 2022 Jan 11;5(1):16. Github page for scripts / Media highlight Description: To better understand the transcriptional activities of the UPR master regulators, bZIP28 and bZIP60, I generated time-series transcriptome datasets and performed coexpression analyses, which allowed me to pinpoint downstream genes for functional characterization. |
|
|
Plant Cell Atlas Consortium, Ghosh Jha S, Borowsky AT .... Ko DK ... Rhee SY eLife. 2021 Sep 7;10:e66877. Plant Cell Atlas website Description: Plant Cell Atlas (PCA) colleagues and I reported the activities that we initiated for the plant research community. |
|
|
Angelos E, Ko DK, Zemelis-Durfee S, Brandizzi F Astrobiology. 2021 Mar;21(3):367-380. Media highlight Description: This study focused on investigating transcriptome changes affected by UPR regulators in space. Plants were germinated, grown, and harvested in spaceflight. As a co-author, I performed RNA-seq analyses. |
|
|
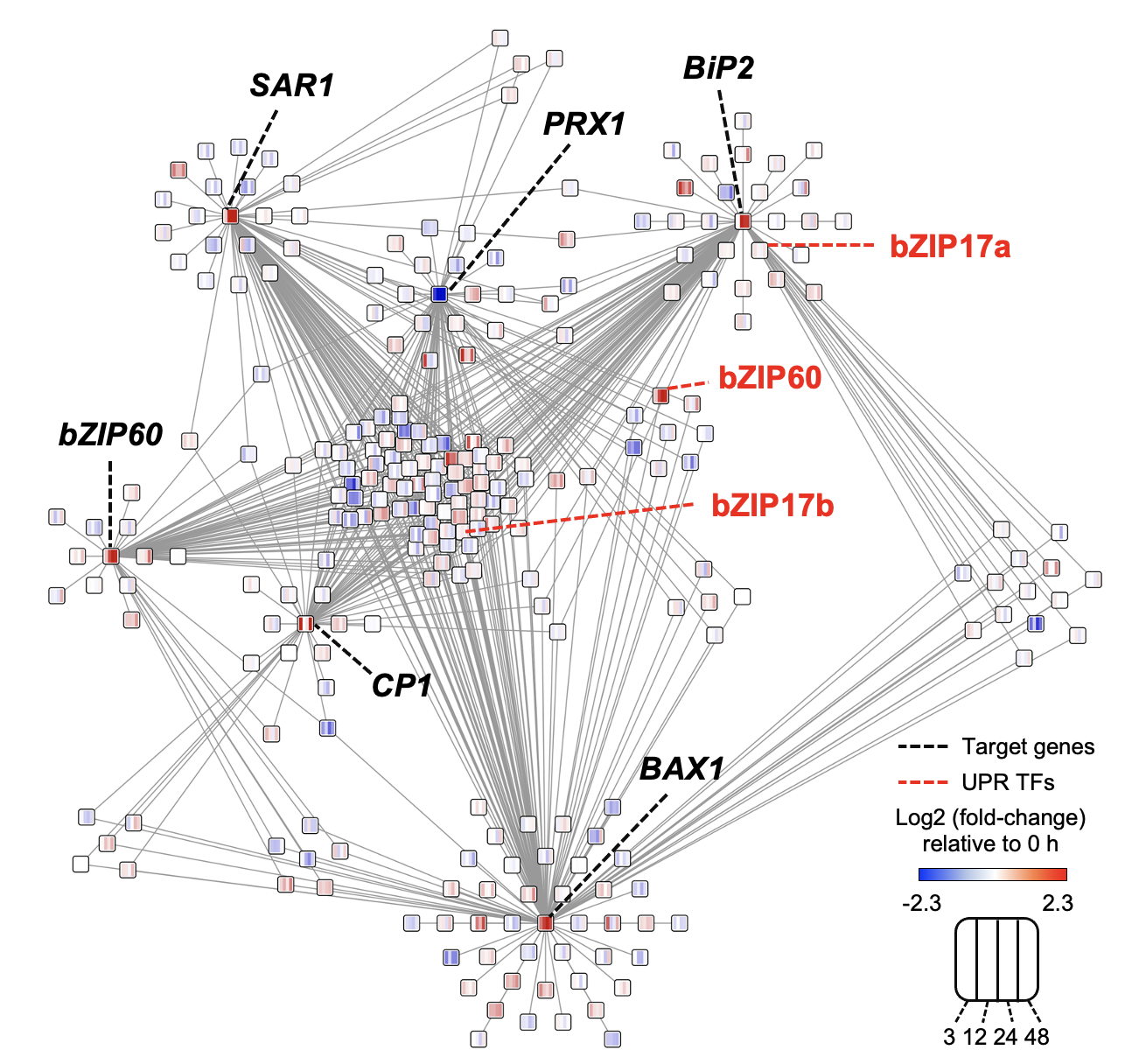
Ko DK, Brandizzi F Plant J. 2021 Jan;105(1):254-270. Media highlight Description: How the UPR is controlled is largely unknown in non-model plant species including maize. To better understand the regulatory landscape of the UPR, I performed TF network analyses based on enhanced Y1H screens for selected UPR marker genes in maize. |
|
|
Rice S, Fryer E, Ghosh Jha S...The Plant Cell Atlas Consortium (including Ko DK) Plant Direct. 00:1–10. Plant Cell Atlas website Description: In this white, Plant Cell Atlas (PCA) colleagues and I report the very first workshop that was successfully held online |
|
|
Ko DK, Brandizzi F Plant J. 2020 Oct;104(2):302-317. Highlighted in the Society for Experimental Biology's Spring Bulletin Description: Understanding gene function is a critical requirement to advance knowledge of the principles underpinning fundamental and applied plant biology. In this review, we describe predictive analyses, their strengths and pitfalls that are fast-forwarding our understanding of gene regulation and function in plants. |
|
|
Pastor-Cantizano N, Ko DK, Angelos E, Pu Y, Brandizzi F. Trends Biochem Sci. 2020 Feb;45(2):123-136. On The Cover Description: In recent years, the field of plant UPR has substantially advanced, revealing new regulators and mechanisms. My colleagues and I provide recent updates on the new development in the plant UPR to the research community. |
|
|
Nadakuduti SS, Starker CG, Ko DK, Jayakody TB, Buell CR, Voytas DF, Douches DS. Front Plant Sci. 2019 Feb 8;10:110. Description: We compare three existing methods for detection of gene editing activity in potato protoplasts. In this collaborative project, I performed amplicon-seq analyses as a co-author. |
|
|
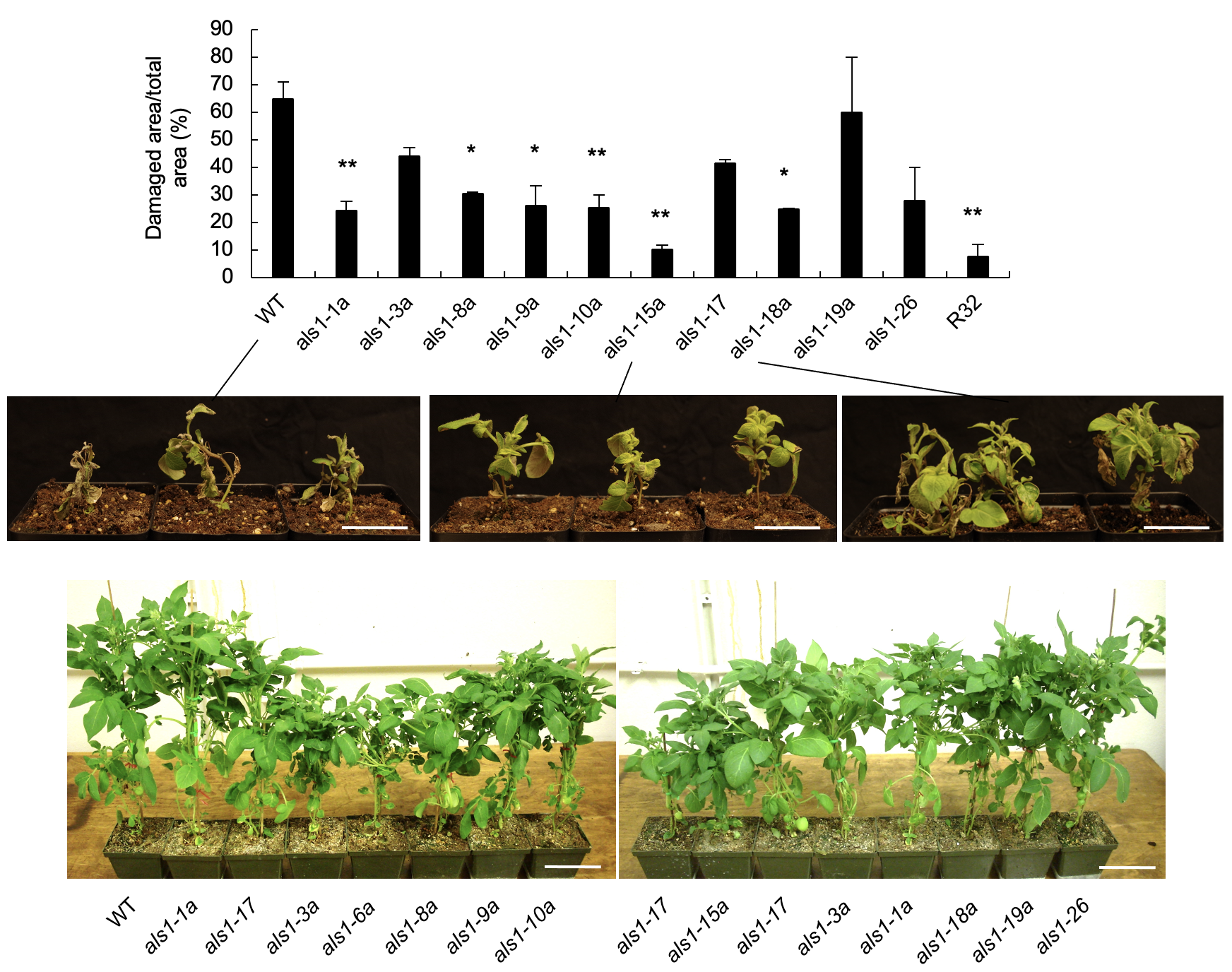
Ko DK, Nadakuduti SS, Douches DS, Buell CR. PLoS One. 2018 Nov 8;13(11):e0206055. Description: We describe the impact of Agrobacterium-mediated transformation of a mutated acetolactate synthase gene on the transcriptome of potato, highlight the extent of positional effect of transgene insertion and that one component of somaclonal variation is due to altered expression of transcription factors and their downstream targets. |
|
|
|
Ko DK*, Rohozinski D*, Song Q, Taylor SH, Juenger TE, Harmon FG, *Chen ZJ. *These authors contributed equally to this work PLoS Genet. 2016 Jul 28;12(7):e1006197. Description: Heterosis, or hybrid vigor, has been widely used in agriculture for more than a century. Despite extensive investigation and various models proposed, the molecular basis for heterosis remains largely elusive. I led this collaborative project to identify and characterize the molecular link of growth heterosis and the circadian clock. We report that higher carbon fixation and starch accumulation in maize hybrids than in the parents are associated with differences in the diurnal regulation of gene expression. |
|
Shi X, Zhang C, Ko DK, Chen ZJ. Mol Biol Evol. 2015 Sep;32(9):2351-66. Description: We investigate allelic expression novelties using a series of newly synthesized Arabidopsis tetraploids that contain one, two, three, and four genomes of A. thaliana or A. arenosa, as well as reciprocal crosses containing the same nuclear genomes but different cytoplasms. |
|
|
Ko DK, Lee MO, Hahn JS, Kim BG, Hong CB. J Plant Physiol. 2009 Jul 1;166(10):1090-100.. Description: In my MS research, I identified and characterized a flooding stress-responsive transcription factor gene of which expression was not only responsive to flooding stress but also under the circadian clock regulation in Nicotiana. It demonstrated potential crosstalk between the clock and abiotic stress responses. |
|
|